Every organism has its own family tree. And like any family tree, an organism’s family tree is more interesting if it’s both complete and richly detailed. That is, each member of the family tree should be displayed in their proper place along with some biographical information. In the case of an organism—a mouse, for example—the family tree’s members are individual cells, and the biographical information consists of gene expression profiles.
If comprehensive whole-organism family trees could be assembled, researchers would learn a lot about development, aging, and disease. Unfortunately, family trees that trace tissue or organism development have been limited to small groups of cells or rendered vaguely suspect, due to distortions cause by intrusive cell assessment techniques.
The good news is that a new technology has been developed that can serve as a sort of ancestry.com for an organism’s cells. That is, it promises to couple cell ancestry information with detailed molecular readouts, such as transcriptional signatures.
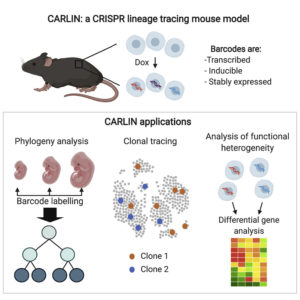
The technology, which is called CRISPR Array Repair Lineage tracing (CARLIN), was developed by scientists at the Stem Cell Research program at Boston Children’s Hospital and Dana-Farber Cancer Institute/Harvard Medical School. It can be used to track every cell in the body, from the embryonic stage until adulthood.
Using a “barcoding” technique and CRISPR gene-editing technology, CARLIN can identify different cell types as they emerge and what genes each is turning on. Details about CARLIN emerged in the journal Cell, in an article titled, “An Engineered CRISPR-Cas9 Mouse Line for Simultaneous Readout of Lineage Histories and Gene Expression Profiles in Single Cells.”
“This model exploits CRISPR technology to generate up to 44,000 transcribed barcodes in an inducible fashion at any point during development or adulthood, is compatible with sequential barcoding, and is fully genetically defined,” the article’s authors wrote. “We have used CARLIN to identify intrinsic biases in the activity of fetal liver hematopoietic stem cell (HSC) clones and to uncover a previously unappreciated clonal bottleneck in the response of HSCs to injury.”
“The dream that many developmental biologists have had for decades is a way to reconstruct every single cell lineage, cell by cell, as an embryo develops, or as a tissue is built up,” said Fernando Camargo, PhD, a senior investigator in the Stem Cell Research program and co-senior author on the paper with Sahand Hormoz, PhD, researcher, Dana-Farber Cancer Institute and assistant professor of systems biology, Harvard Medical School. “We could use this mouse model to follow its entire development.”
Camargo, Hormoz, and co-first authors for their respective labs—Sarah Bowling, PhD, and Duluxan Sritharan—created the mouse model using a method they call CRISPR Array Repair Lineage tracing, or CARLIN. The model can reveal cell lineages—the “family tree” in which parent cells create different types of daughter cells—as well as what genes are turned on or off in every cell over time.
Previously, scientists have only been able to trace small groups of cells in mice using dyes or fluorescent markers. Tags or barcodes have also been used, but previous approaches required prior knowledge of markers to isolate different cell types, or required time-consuming extraction and manipulation of cells, which could affect their properties. The advent of CRISPR has enabled researchers to barcode cells without perturbing the cells and to follow the lineage of thousands of cells simultaneously.
Using an inducible form of CRISPR, the researchers can create up to 44,000 different identifying barcodes at any time point in a mouse’s lifespan. The scientists can then read out the barcodes using another technology called single-cell RNA sequencing, enabling collection of information on thousands of genes that are turned on in each barcoded cell. This, in turn, provides information about the cells’ identity and function.
As a test case, the researchers used the new system to reveal unknown aspects of blood development during embryonic development, and to observe the dynamics of blood replenishment after chemotherapy in adult mice.
But the researchers believe their system could also be used to understand the changes in cellular lineage trees during disease and aging. Additionally, the system could be used to record the response to environmental stimuli like pathogen exposure and nutrient intake.
“Being able to create single-cell lineage maps of mammalian tissues is unprecedented,” said Camargo, who is also a member of the Harvard Stem Cell Institute. “Besides its many applications to studying developmental biology, our model will provide important insight on the cell types and hierarchies that are affected as organisms respond to injury and disease.”